FoodOn’s OWL file format and its attendant RDF graph representation contain fairly easy to query (with SPARQL) class-subclass (parent-child), annotation (label, definition, synonym, comment), and object properties (‘has ingredient’, ‘has food product analog’, etc.) about food products and categories. Querying some of the more complex information stored in logical statements – like equivalency and subclass expressions about a harvested food item’s plant or animal part and the taxonomy of the organism it came from – is more challenging unless we first transform the owl file to make explicit the simpler facts. Consider what “apple (whole)”(FOODON:00002473) looks like in Protege:
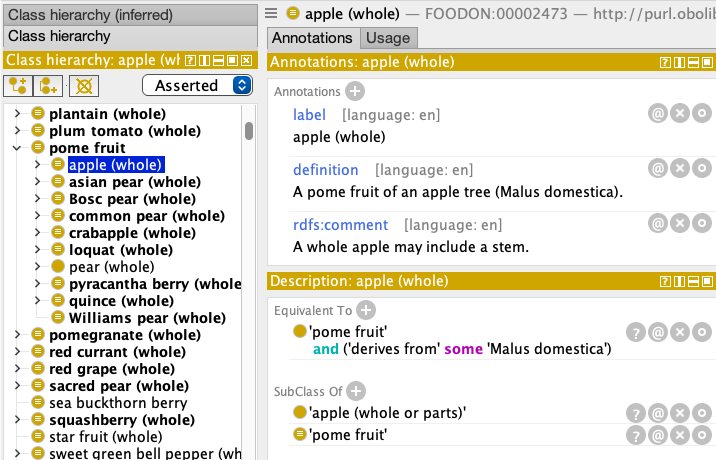
The “Equivalent To” expression seems fairly straightforward in its logical statement, but when it comes to querying anything with brackets or logical keywords like “and”, “or”, “some”, “only”, the representation in an OWL file (like src/ontology/imports/robot_plant_parts.owl) and its RDF graph equivalent gets gnarly:
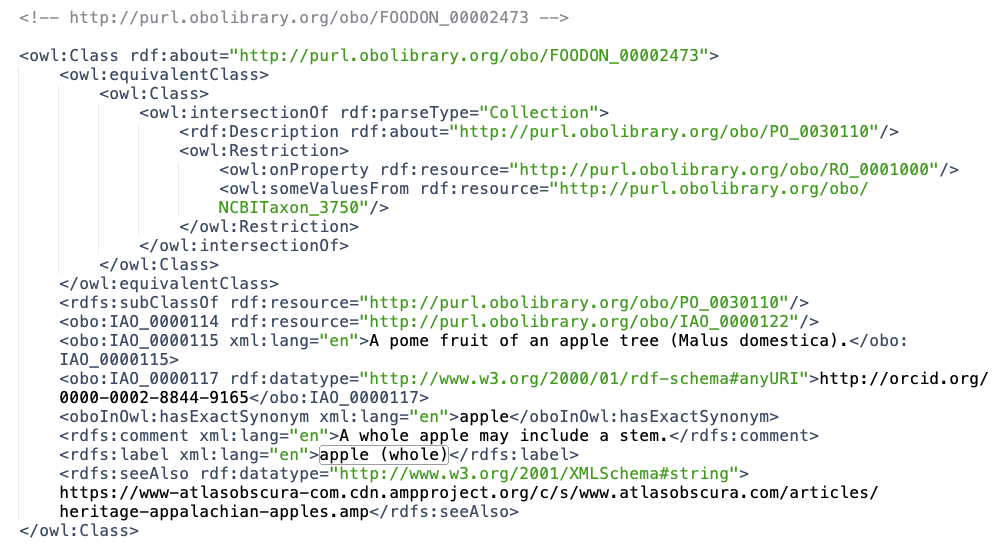
“‘pome fruit’ and (‘derives from’ some ‘Malus domestica’)”
The fact that an apple is a pome fruit, and the fact that it derives from some Malus domestica is buried inside the complex equivalency statement, <owl:equivalentClass>….</owl:equivalentClass> which is designed for categorization of entities bearing those facts. To get these facts exposed so they are easier to query, we need to run a robot command to unpack the equivalency statement into simpler SubClassOf (i.e. necessary) conditions.
robot relax --input imports/robot_plant_parts.owl --output reasoned.owl
Now, opening “reasoned.owl” and finding the same “apple (whole)” entry above, you should see this extra set of clauses:
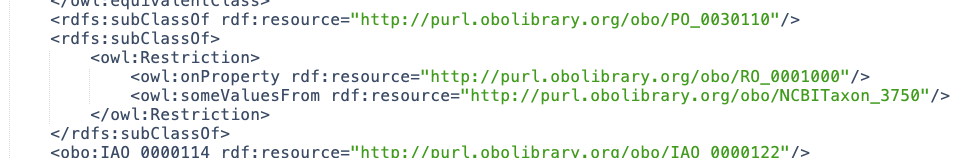
Here is a simpler query to fetch the anatomical part, “pome” (PO:0030110), and the taxonomy:
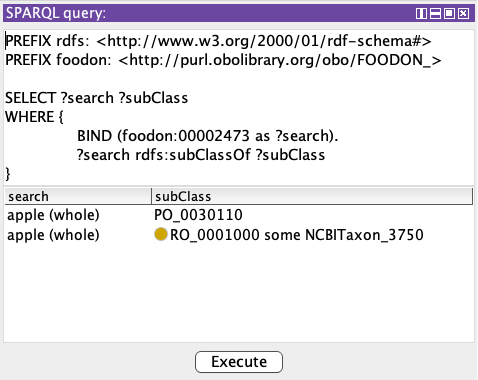
Admittedly that second result’s “subClass” value, the taxonomy link, is actually still in a blank or anonymous node which the SPARQL result display is hiding the complexity of. We need a larger query that taps into the <owl:Restriction>…</owl:Restriction> expression listed above. Below the searched-for term is shown, along with linked parent or taxon, and the “type” column gives a label to the kind of relationship found.
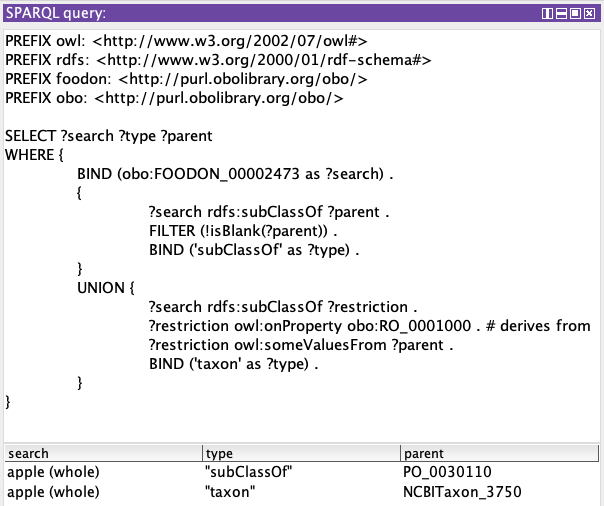
Also note that by dropping the ?search BIND statement, ?search could be loosened up to report on any class/subclass relation in the given ontology.
Here is the text of the query to play with if desired:
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX foodon: <http://purl.obolibrary.org/obo/>
PREFIX obo: <http://purl.obolibrary.org/obo/>
SELECT ?search ?type ?parent
WHERE {
BIND (obo:FOODON_00002473 as ?search) .
{
?search rdfs:subClassOf ?parent .
FILTER (!isBlank(?parent)) .
BIND ('subClassOf' as ?type) .
}
UNION {
?search rdfs:subClassOf ?restriction .
?restriction owl:onProperty obo:RO_0001000 . # derives from
?restriction owl:someValuesFrom ?parent .
BIND ('taxon' as ?type) .
}
}