Use of FoodOn in other projects is listed here; let us know of your use and we will include it! For technical advice on how to incorporate FoodOn into your project, see: https://foodon.org/reuse-technical.
The Canadian Antimicrobial Resistance Genomics Research and Development Initiative (AMR-GRDI)
The GRDI project uses a genomics-based approach to understand how food production and the environment contribute to the development of antimicrobial resistance of human health concern, and to explore strategies for reducing antimicrobial resistance using a One Health approach. The AMR-GRDI is a component of the Federal Action Plan for Antimicrobial Resistance and Use in Canada, and involves data streams from federal departments and agencies spanning human health, agriculture, the environment, and food regulation, and so FoodOn is a key ontology within the sample descriptors of this project.
https://grdi.canada.ca/en/projects/antimicrobial-resistance-amr-project
USDA Agricultural Research Service FoodData Central Data Portal:
The FoodData Central portal is USDA’s comprehensive source of food composition (nutritional) data with multiple distinct data types drawn from the agency’s SR Legacy database, Foundation Foods, Experimental Foods, Branded Foods, and FNDDS survey foods. FoodOn food product categories are offered for Experimental Foods, and are being added to the SR Legacy single ingredient food sample descriptions, with the intent to provide the foundation upon which to calculate nutritional outcomes of food processes applied to such foods.
fdc.nal.usda.gov
FDA GenomeTrackr Network
To quote: “The GenomeTrakr network is the first distributed network of laboratories to utilize whole genome sequencing for pathogen identification. It consists of public health and university laboratories that collect and share genomic and geographic data from foodborne pathogens. The data, which are housed in public databases at the National Center for Biotechnology Information (NCBI), can be accessed by researchers and public health officials for real time comparison and analysis that can speed foodborne illness outbreak investigations and reduce foodborne illnesses and deaths.” GenomeTrakr uses FoodOn food product classes to describe food samples.
https://www.fda.gov/food/whole-genome-sequencing-wgs-program/genometrakr-network
Periodic Table Of Food Initiative
The Periodic Table Of Food Initiative (PTFI) is Rockefeller-funded web portal and database of plant and animal food samples and their molecular constituents, collected from participating global laboratories according to the same standardized analytic methods, and using ontologies including FoodOn and NCBITaxon to describe each sample. Launched in April 2024.
https://foodperiodictable.org/
WikiFCD
WikiFCD is an open source knowledge base of structured data related to food items and their nutritional content drawn from national food composition databases (FCDs) integrated from across the globe. A subset of the FoodOn ontology food product hierarchy was imported into the WikiFCD knowledge base as a cross reference to FCD foods, along with NCBI taxon identifiers. Reusing FoodOn benefits WikiFCD by allowing us to leverage the food item groupings that FoodOn contains, which yeilds a more powerful querying of database content via the WikiFCD SPARQL endpoint.
https://wikifcd.wikibase.cloud/wiki/Main_Page and paper
CGIAR
CGIAR is an international organization supporting agricultural research and data exchange within and across continents. To quote “In the Systems Transformation Action Area, CGIAR commits to forge, with partners, ambitious new multi-sectoral policies and strategies for food, land, and water systems transformation in 50 countries across six regions, via a set of ambitious Initiatives …” CGIAR has recommended the use of ontologies such as FoodOn and AGRO in describing agricultural practices and products.
https://www.cgiar.org/ and https://bigdata.cgiar.org/blog-post/agricultural-ontologies-in-use-foodon-a-farm-to-fork-food-description-ontology/
Comfocus
COMFOCUS, a European Horizons funded project to provide the academic and private research community with easy virtual and transnational access to high quality services and resources in the consumer behaviour research area, includes a data harmonization component for developing measurement standards and survey tools, along with recommended reuse of ontologies such as FoodOn and ONS.
https://comfocus.eu/ and https://comfocus.eu/wp-content/uploads/2023/11/Report-on-basic-ontology-in-food-consumer-science.pdf
FoodKG
FoodKG (https://foodkg.github.io/), a knowledge graph representing over 1 million recipes which is constructed with an ontology combining FoodOn, CHEBI and a few other resources like the USDA Nutrient Database, enables querying of recipes by ingredient, cook time, course type, and meal type. Other applications such as natural language querying are explored in the project, which is supported by the Health Empowerment by Analytics Learning and Semantics (HEALS) project.
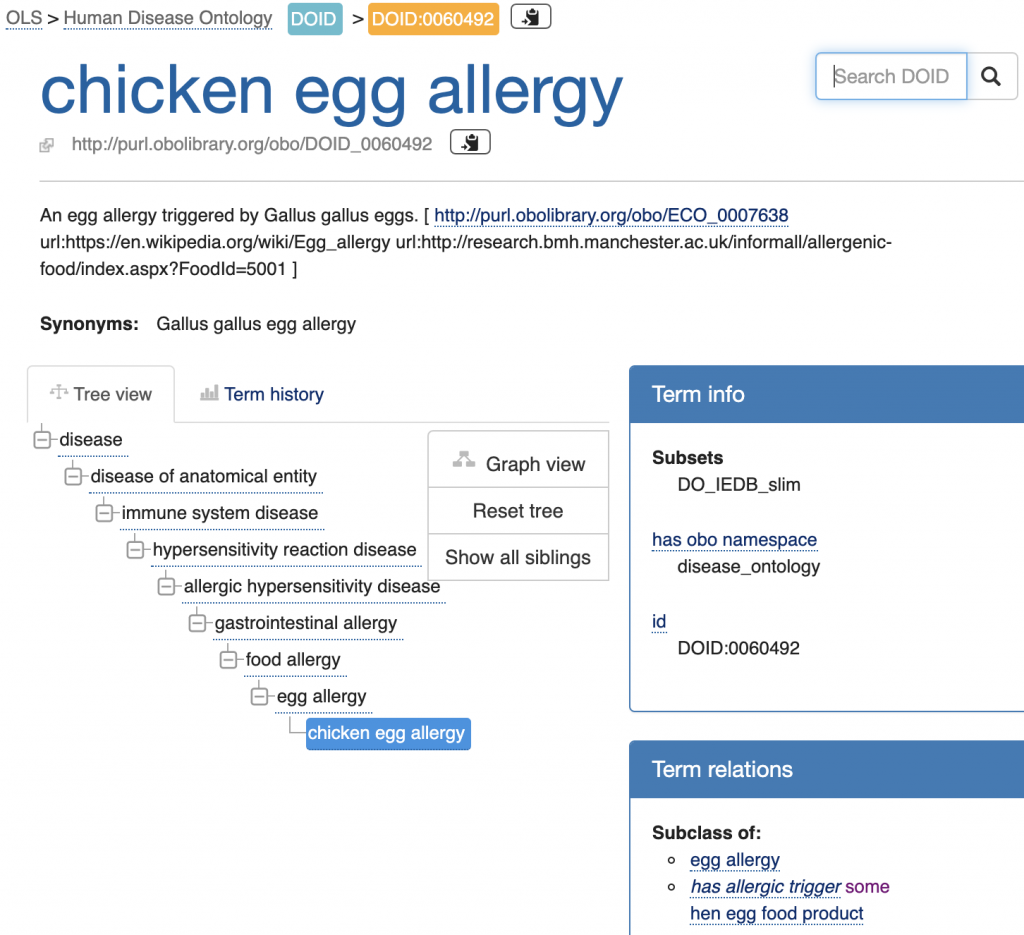
Human Disease Ontology (DOID)
In connection with Immune Epitope Database (IEDB) content related to food allergens (http://www.iedb.org/) FoodOn product classes are being referenced via the “has allergic trigger” relation.
Food-Biomarker Ontology (FOBI)
FOBI describes food products and their associated metabolite entities in a hierarchical way, drawing heavily on FoodOn, CHEBI, and its own biomarker-specific classes. An extract from the recently launched FOBI poster:
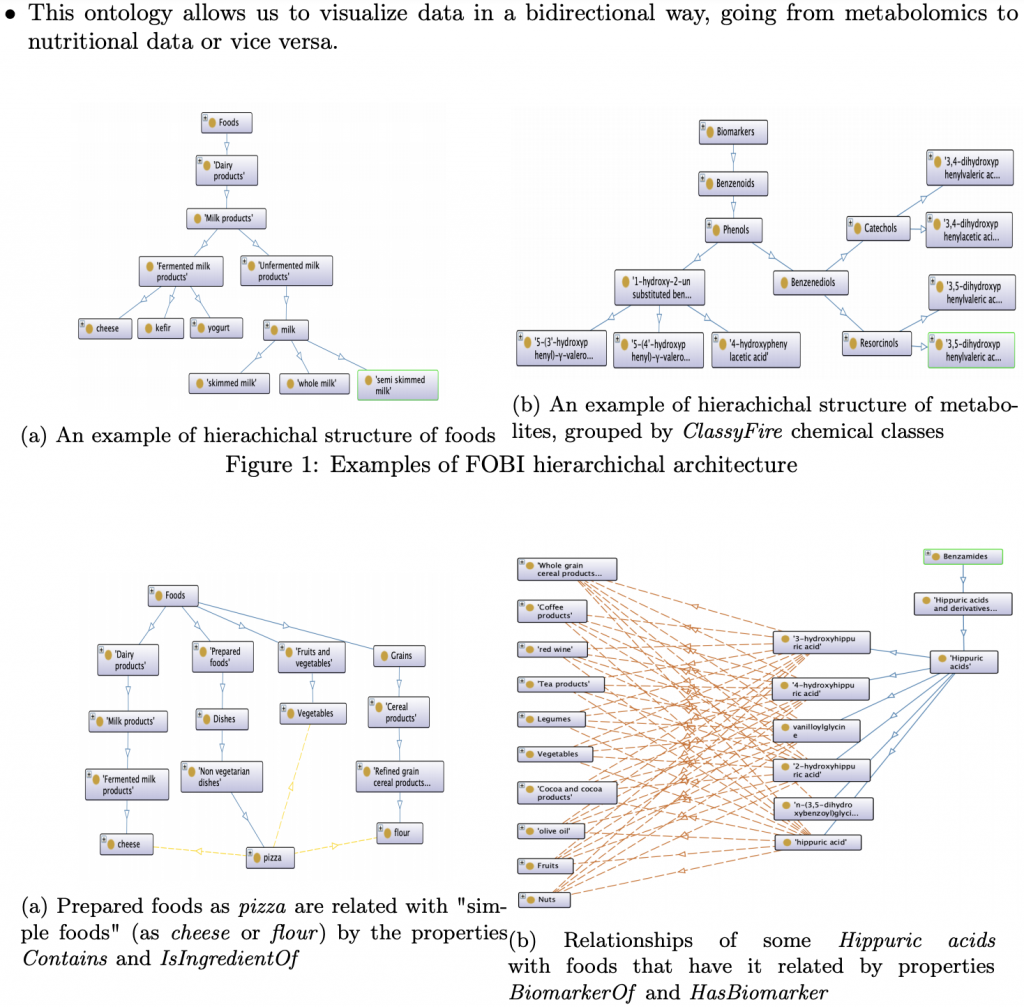
For more information, see: https://pcastellanoescuder.github.io/talk/fobi_bioinformatics and https://github.com/pcastellanoescuder/FoodBiomarkerOntology and https://grbio.upc.edu/en/shared/20190409_TALKPCastellano.pdf
Ontology for Nutritional Studies (ONS)
Quoting fromONS: “An ontology for a standardized description of interventions and observational studies in nutrition” (https://www.ncbi.nlm.nih.gov/pubmed/29736190), “The ONS was developed within the European Nutritional Phenotype Assessment and Data Sharing Initiative (ENPADASI) consortium, which joins scientists from 51 research centers in nine countries of Europe with the common effort to handle and make available big nutritional data through the open access nutritional database Data Sharing In Nutrition (DASH-IN) […] The development of this infrastructure requires an ontology to harmonize biochemical, genetic, clinical, and nutritional concepts typically found in intervention and observational studies.” A diagram from the paper of ONS illustrates the interplay of a number of OBOFoundry related ontologies, including FoodON.
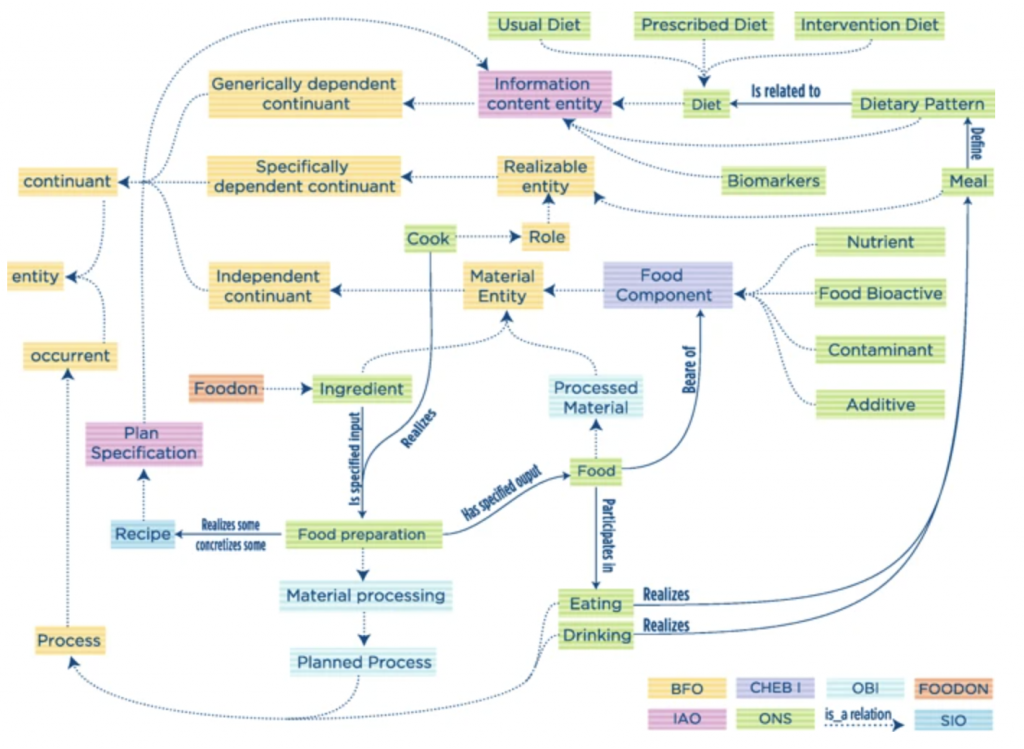
https://github.com/enpadasi/Ontology-for-Nutritional-Studies/
Food Interactions with Drugs Evidence Ontology (FIDEO)
Built as part of the MIAM – Diseases, Food/Drug Interaction project, FIDEO was developed to support food-drug interactions automatically extracted from scientific literature. It draws upon BFO, DIDEO, FoodOn, and ChEBi ontologies.