Use of FoodOn in other projects is listed here; let us know of your use and we will include it! For technical advice on how to incorporate FoodOn into your project, see: https://foodon.org/reuse-technical.
FoodKG
FoodKG (https://foodkg.github.io/), a knowledge graph representing over 1 million recipes which is constructed with an ontology combining FoodOn, CHEBI and a few other resources like the USDA Nutrient Database, enables querying of recipes by ingredient, cook time, course type, and meal type. Other applications such as natural language querying are explored in the project, which is supported by the Health Empowerment by Analytics Learning and Semantics (HEALS) project.
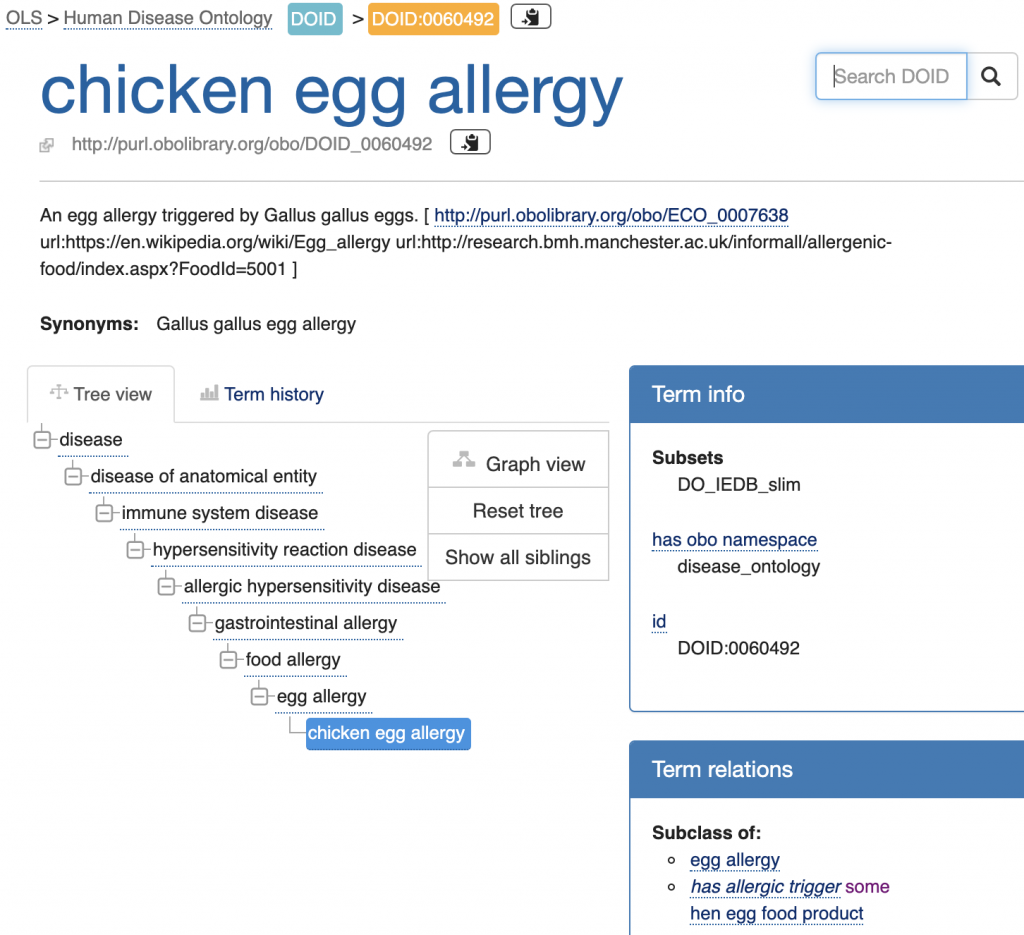
Human Disease Ontology (DOID)
In connection with Immune Epitope Database (IEDB) content related to food allergens (http://www.iedb.org/) FoodOn product classes are being referenced via the “has allergic trigger” relation.
Food-Biomarker Ontology (FOBI)
FOBI describes food products and their associated metabolite entities in a hierarchical way, drawing heavily on FoodOn, CHEBI, and its own biomarker-specific classes. An extract from the recently launched FOBI poster:
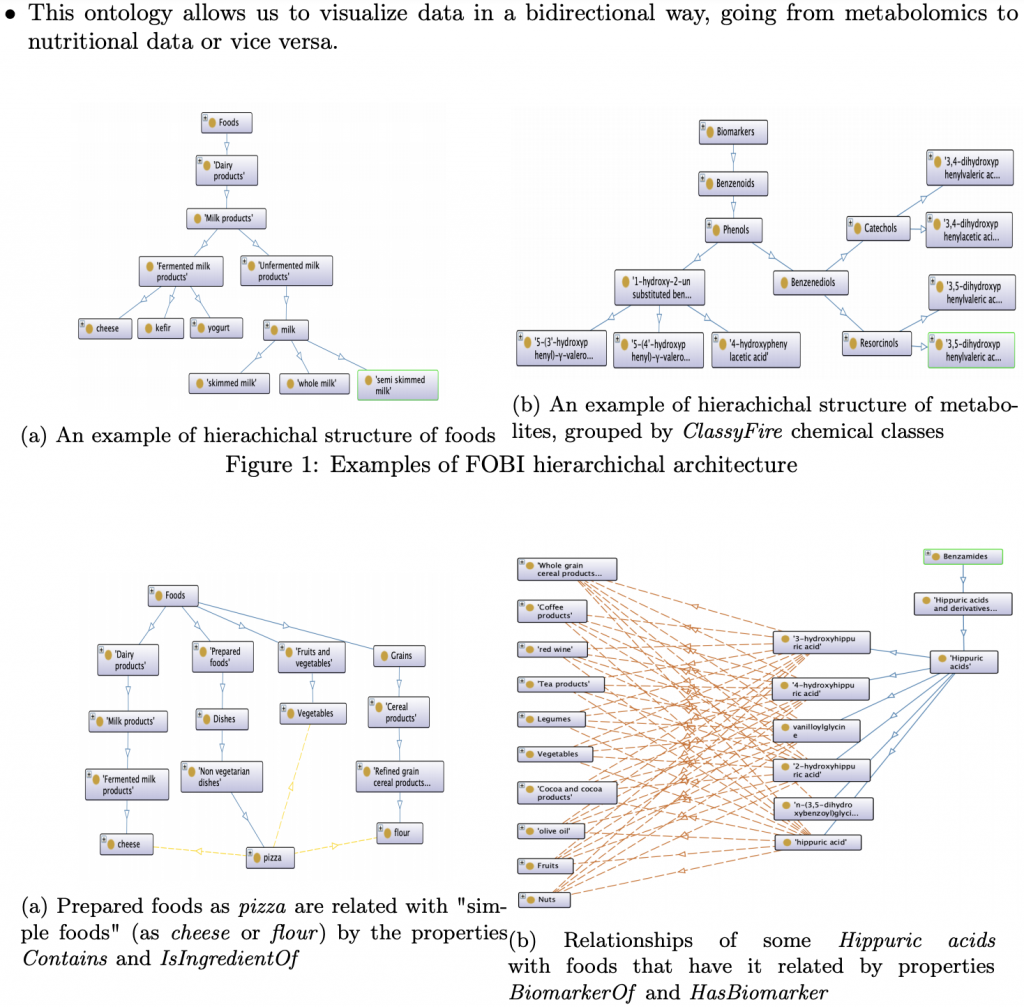
For more information, see: https://pcastellanoescuder.github.io/talk/fobi_bioinformatics and https://github.com/pcastellanoescuder/FoodBiomarkerOntology and https://grbio.upc.edu/en/shared/20190409_TALKPCastellano.pdf
Ontology for Nutritional Studies (ONS)
Quoting fromONS: an ontology for a standardized description of interventions and observational studies in nutrition (https://www.ncbi.nlm.nih.gov/pubmed/29736190), “The ONS was developed within the European Nutritional Phenotype Assessment and Data Sharing Initiative (ENPADASI) consortium, which joins scientists from 51 research centers in nine countries of Europe with the common effort to handle and make available big nutritional data through the open access nutritional database Data Sharing In Nutrition (DASH-IN) […] The development of this infrastructure requires an ontology to harmonize biochemical, genetic, clinical, and nutritional concepts typically found in intervention and observational studies.” A diagram from the paper of ONS illustrates the interplay of a number of OBOFoundry related ontologies, including FoodON.
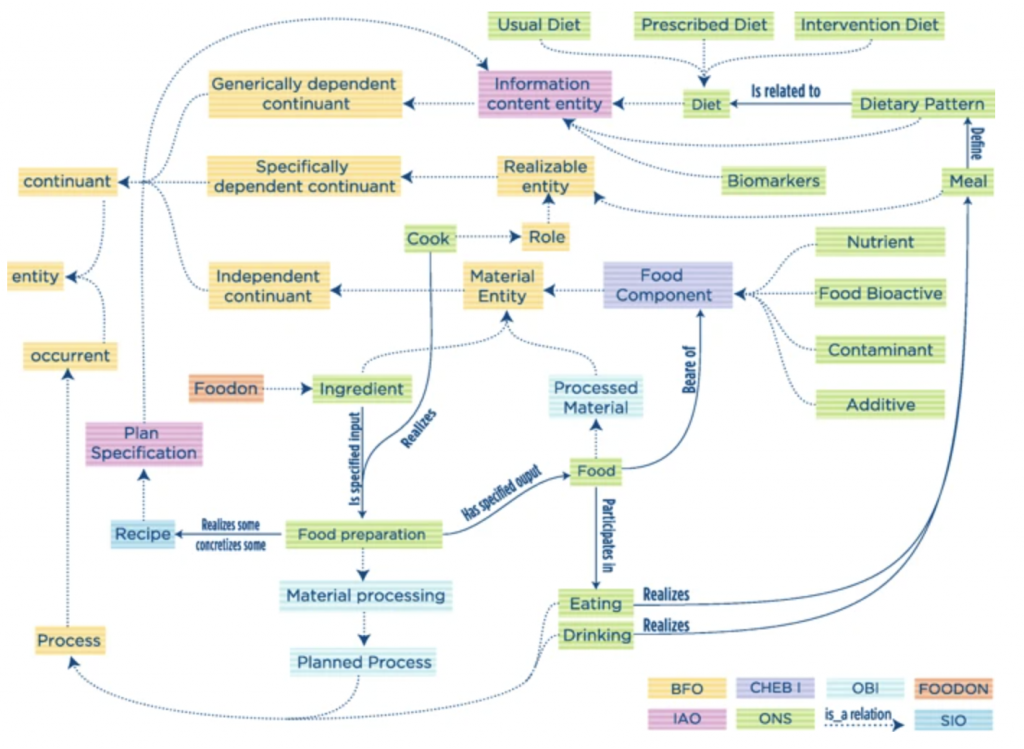
http://bioportal.bioontology.org/ontologies/ONS
Food Interactions with Drugs Evidence Ontology (FIDEO)
Built as part of the MIAM – Diseases, Food/Drug Interaction project, FIDEO was developed to support food-drug interactions automatically extracted from scientific literature. It draws upon BFO, DIDEO, FoodOn, and ChEBi ontologies.